The genetics and genomics of thoracic aortic disease
Introduction
Thoracic aortic aneurysms (TAA) and aortic dissections (TAAD) are responsible for 15,000 deaths in the United States annually (1). Despite progressive dilatation, aortic aneurysms usually remain asymptomatic until dissection or rupture occurs. While there are highly effective prophylactic surgical interventions, their implementation is hampered by the difficulties in identifying at-risk subjects. Indeed, there are no high-yield risk factors that can be used for screening the general population. Identification of the underlying genetic basis of aortic aneurysms should lead to better screening, early intervention, and better clinical outcomes.
Thoracic aortic aneurysms are divided into two broad categories: syndromic (associated with abnormalities of other organ systems) and non-syndromic (with manifestations restricted to the aorta) (2).
Syndromic aortic aneurysms
Hippocrates first described a clinical syndrome consistent with Ehler-Danlos and its accompanying easy bruising and bleeding as far back as 400 BC. Writing in his “Airs, Waters, and Places”, he noted that Nomads and Scythians had lax joints and easy bruising. The Danish dermatologist Ehler in 1901 and the French physician Danlos in 1908 then refined the clinical description of the disorder. Several years earlier, Antoine Marfan, a French pediatrician, had described a hereditary connective tissue disorder which came to bear his name. His report was made in 1896 in the Bulletin of the Medical Society of Paris and described a five-year-old girl with long limbs and digits (3). It was not until over 50 years later that the syndrome was fully described, including the involvement of aneurysms of the ascending aorta. In 2006, Loeys and Dietz described the syndrome of early, malignant arterial dilatations and unique facial features which characterize the syndrome that bears their names (4). These were the first clinical reports of recognizing the genetically heterogenous nature of thoracic aorta aneurysms.
Syndromic aortic aneurysms occur in patients with: Marfan syndrome (MFS), Loeys-Dietz syndrome (LDS), aneurysm osteoarthritis syndrome (AOS), arterial tortuosity syndrome (ATS), Ehlers-Danlos Syndrome (EDS) (2,5) and TGFβ mutations. Many of the syndromic aneurysms can be diagnosed by their characteristic dysmorphic features and gene testing (Tables 1,2).

Full table

Full table
Marfan syndrome
Marfan syndrome (MFS) is the best known aortic aneurysm syndrome and is caused by heterozygous mutations in the FBN1 gene. It occurs worldwide with an estimated incidence of 1 in 5,000 individuals and affects both sexes equally. Clinically MFS is a multisystem disease affecting the aorta (aortic aneurysm/dissection), the heart (mitral/aortic valve insufficiency), the eyes (ectopia lentis), and the musculoskeletal system (overgrowth). The diagnosis of Marfan syndrome is made based on the revised Ghent Nosology, which incorporates clinical criteria (based on a point score system), family history, and molecular genetic testing of the FBN1 gene (Table 3). Marfan syndrome follows an autosomal dominant pattern of inheritance with high penetrance (the probability of manifesting a disease) and significant inter/intra-familial variability in disease expression (6). In the majority of MFS patients, the disease is caused by a mutation in FBN1 gene on chromosome 15q21.1, which encodes an extracellular matrix protein, fibrillin-1 (7). Over 600 FBN1 gene mutations have been identified to date, and the detection rate for the FBN1 mutations by DNA sequencing ranges from 70-93% in patients who meet the clinical diagnostic criteria (8). Various types of mutations in the FBN1 gene have been identified, including nonsense, deletion/insertion, splice-site mutations, and missense mutations (9) with some genotype-phenotype correlations. The most severe forms of Marfan syndrome are associated with FBN1 mutations between exons 24-32 causing in-frame loss or gain of central coding sequences. Mutations causing premature stop codons result in rapid degradation of mutant transcripts and usually present with a milder phenotype (10).

Full table
Loeys-Dietz syndrome
A subset of patients previously thought to have Marfan syndrome do not have identifiable mutations in FBN1. Most of these patients are now classified as having Loeys-Dietz syndrome (LDS) (4,11). LDS is an autosomal dominant disorder of connective tissue with multisystem involvement (2). Patients with typical dysmorphic features (hypertelorism, craniosynostoses, cleft palate) are classified as LDS type 1; those with mild dysmorphic features (only hypertelorism) without other dysmorphic findings but with velvety, translucent skin, easy bruising, and atrophic scars are classified as LDS type 2 (12). The natural history of both types of LDS is marked by early and rapid onset of thoracic aortic aneurysms (originating at the level of the sinuses of Valsalva) and death at an early age (mean age reported at 26.1 years) (12).
LDS is caused by a heterozygous mutation in one of the transforming growth factor beta receptor genes (TGFβR1 or TGFβR2). TGFβR2 mutations on chromosome 3 (3p24.1) account for the majority of LDS mutations (75%), and TGFβR1 mutations on chromosome 9 (9q22.33) account for the remaining 25% (12). Direct sequencing of both TGFβR1 and TGFβR2 allows for the identification of a causal mutation in more than 95% of individuals. Although most LDS mutations are missense, other types of mutations have been seen including: splice-site, nonsense, deletions, and insertions. In LDS, de novo mutations are found in 75% of patients. Both TGFβR1 and TGFβR2 gene mutations are thought to result in the overall up-regulation of TGFβ signaling. The classical TGFβ signaling pathway involving the Smad-mediated cascade has been characterized by signals that not only induce extracellular matrix (ECM) deposition but also matrix degradation. This implicates the pathway as being critical to the structure and composition of ECM (13). Enhanced TGFβ signaling with downstream canonical pSmad2 upregulation has been described as the predominant mechanism in both Marfan and Loeys-Dietz aneurysm formation (11).
Aneurysm osteoarthritis syndrome
Aneurysm osteoarthritis syndrome (AOS) is a newly described autosomal dominant syndrome with variable expression which presents with aortic aneurysms in the setting of atypical osteoarthritis and dysmorphic features. The frequency of AOS is 2% among TAA patients. AOS is caused by mutations in the Smad3 gene on chromosome 15 (15q22.33), which encodes a protein critical for cellular signaling downstream of the TGFβ receptors (14). Mutations in Smad3 result in aortic aneurysms, dissections, arterial tortuosity, early onset osteoarthritis, and cutaneous anomalies. In one series, aortic aneurysms were present in 71% of patients with Smad3 mutations, mainly at the level of the sinus of Valsalva but also affecting the abdominal aorta and/or other arteries such as the splenic, common iliac, mesenteric, renal, vertebral, and pulmonary arteries. The mean age of death, due to aortic dissection, has been reported at 54±15 years and occurred at mildly increased aortic diameters (4.0-6.3 cm) (15). Arterial tortuosity was diagnosed in 48% of patients. Velvety skin, striae, and umbilical/inguinal hernias are common and joint abnormalities; osteochondritis dissecans (OCD), meniscal abnormalities, intervertebral disc degeneration, and early-onset osteoarthritis have been reported in all cases of AOS (16). Cerebrovascular abnormalities were also noted to be common, and in 18% of patients concentric left ventricular hypertrophy (LVH) was observed. Importantly, LVH was not a consequence of hypertension as most patients were normotensive without treatment (17).
Arterial tortuosity syndrome
Arterial tortuosity syndrome (ATS) is an autosomal recessive disorder caused by mutations in the SLC2A10 gene on chromosome 20 (20q13.1), which encodes the facilitative glucose transporter GLUT10 (18-20). GLUT10 is localized to the gene promoter region of decorin, a natural inhibitor of TGFβ. Mutations in the SLC2A10 gene are thought to result in down-regulation of decorin and thereby up-regulation of TGFβ signaling. The mutations which cause this syndrome are loss-of-function mutations. The clinical spectrum of ATS includes: arachnodactyly, joint laxity or contractions, hypertelorism, cleft palate, bifid uvula, micrognathia, down-slanting palpebral fissures, blepharophimosis and arterial tortuosity with aneurysm formation (18).
Ehlers-Danlos Syndrome Type IV
Ehlers-Danlos Syndrome Type IV (EDS IV) is an autosomal dominant disorder with a prevalence of 1:10,000-25,000 in the US (21), caused by highly penetrant mutations in COL3A1 (2q32.2). EDS IV is characterized by cutaneous findings (thin translucent skin, easy bruising) and arterial, intestinal, or uterine rupture. In childhood, inguinal hernias, pneumothoraces, and recurrent joint and hip dislocations are common. Vascular aneurysms/dissections or gastrointestinal perforation constitutes the presenting signs in a majority of adults with EDS IV. The average age for the first major arterial or gastrointestinal complication is 23 years. Among patients with EDS IV ascertained because of a medical complication associated with the disorder, in 25% the complication was present by age 20 and in 80% by age 40 (22). The median age of death in patients with EDS IV is 48 years. The diagnosis of EDS IV is made on clinical grounds (using major and minor diagnostic criteria) and is confirmed by genetic or protein-based testing (23). Surgical intervention is notoriously dangerous in EDS IV due to excess vessel fragility.
To date, at least 700 mutations in COL3A1 have been reported. Using direct sequence analysis 98% of individuals with a clinical diagnosis of EDS IV are shown to have a mutation in COL3A1, with 50% of EDS IV patients having a de novo mutation. The majority of mutations are missense mutations resulting in an amino acid substitution for glycine residues in the [Gly-X-Y]343 triplets of the triple helical domain or splice-site mutations and mutations resulting in mRNA instability. A small proportion of patients (2-3%) carry a genomic deletion (23). Biochemical (protein-based) testing on cultured cells from a skin biopsy is recommended when a mutation is not identified by sequence analysis in a patient with a clinical diagnosis.
TGFβ2 mutation
TGFβ2 (1q41) is the gene most recently identified as a cause of TAA. TGFβ2 proteins are synthesized as peptides that form homodimers cross-linked through disulfide bonds which are cleaved before secretion. Mutations identified (frame-shift and nonsense) are predicted to cause haplo-insufficiency. Clinical features in TGFβ2 mutation carriers are similar to those of other syndromes causing thoracic aortic disease including: cardiovascular [aortic root aneurysm (74%), cerebrovascular disease, and arterial tortuosity], skeletal (pectus deformity, joint hyperflexibility, scoliosis, and arachnodactyly), cutaneous (striae, herniations), and pulmonary (pneumothorax, dural ectasia). Median age of aortic disease presentation is 35 years, with the majority of patients presenting with aneurysms at the sinuses of Valsalva (4.7-5.4 cm). In a small series reported to date, no aortic dissections occurred in individuals younger than 31 years of age (24).
Non-syndromic aortic aneurysms
For non-syndromic inherited aneurysmal disorders, a less heralded breakthrough occurred at Yale University in 1981. One of us (JAE) was a young resident in the audience when the distinguished Professor M. David Tilson, together with his resident protégé Chau Dang, presented at Surgical Grand Rounds their original observation that aneurysmal disease was distinct clinically from occlusive vascular disease—and that abdominal aneurysmal disease tended to run in families. These truly original and iconoclastic observations laid the foundation for much work that was to come (25,26). In 1997 and 1999, Dr. Diana Milewicz in Texas and our team at Yale reported, independently, that non-syndromic thoracic aortic aneurysms tended to run in families. Both teams, remarkably, reported the same likelihood—20%—that any given proband would have a relative with a known aortic aneurysm (27,28). In the years since those observations of familial patterns in thoracic aortic disease, Milewicz and colleagues have gone on to identify via linkage analysis and other genetic techniques, the specific mutations that underlie many cases of familial thoracic aortic aneurysm and dissection (29).
Non-syndromic aortic aneurysms are thus divided into Familial Thoracic Aortic Aneurysms (FTAA), where more than one person in the family is affected, and sporadic TAA, where only a single person in the family is known to have an aneurysm. FTAAs typically present earlier in life than sporadic aneurysms, have a higher annual growth rate, and do not demonstrate association with traditional risk factors for aortic disease such as dyslipidemia or coronary artery disease (28,30). Cases of non-syndromic TAA primarily involve the thoracic aorta, a region from the aortic root to the thoracic descending aorta.
Non-syndromic Familial Thoracic Aortic Aneurysms and Dissections (FTAA and FTAAD) are also genetically heterogeneous. Mutations in five genes (MYH11, TGFβR1, TGFβR2, MYLK, and ACTA2) have thus far been identified and all have autosomal dominant inheritance. Together these account for only 23% of familial non-syndromic cases. Two additional chromosomal regions have been implicated (5q13-14 and 11q23.3-24) but no genes have as yet been identified. Novel genes have been difficult to map by linkage analysis, probably because of incomplete penetrance and/or locus heterogeneity (2,31,32).
MYH11 mutation
Mutations in the MYH11 gene (chromosome 16p13.11) account for 2% of non-syndromic TAADs, and have been often associated with patent ductus arteriosus (PDA) (33). The myosin superfamily is composed of a large class of motor molecules which interact with actin filaments and result in force generation through ATP hydrolysis. The MYH11 gene is composed of 42 exons and encodes MYH11 protein. The vertebrate smooth muscle myosin is composed of a hexameric complex of 2 myosin heavy chains (SM-MHC coded for by MYH11), 2 essential light chains, and 2 regulatory light chains (RLC). Myosin heavy chains consist of a globular head domain composed of ATPase and actin-binding subdomains, a linker region with Ig repeats that bind light chains, and a variable tail region which specializes each myosin to its cellular functions. SM-MHCs coiled-coil domain in the tail region enables the protein to first dimerize and then polymerize to form thick filaments that interact with actin thin filaments. The coiled-coil domain is composed of a 28 residue charge repeat of alternating positive and negative residues. Most of the identified pathogenic MYH11 mutations are located in the coiled-coil domain of the protein (33,34) (Figure 1).
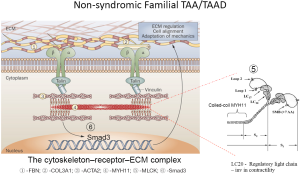
Heterozygous MYH11 mutations result in thoracic aortic aneurysms and are thought to act via a dominant negative mechanism. Mutations in MYH11 identified in FTAAD families are predominantly splice-site mutations resulting in in-frame deletions and missense mutations. Identification of MYH11 mutations responsible for FTAAD is often difficult as in the general population. According to the Exome Rare Variant database, 0.6% of such variants have been identified for MYH11 (
ACTA2 mutation
Mutations in ACTA2 gene are the most common mutations resulting in aortic aneurysms and account for 10-15% of all FTAAD mutations. They are heterozygous mutations which encode the smooth muscle cytoskeletal protein actin alpha 2 (actin α2), which is the vascular smooth muscle specific isoform involved in vascular smooth muscle cell contraction, interacting with the β-myosin heavy chain (encoded by MYH11) (36). The ACTA2 gene is localized to chromosome 10 (10q23.31). Mutations in ACTA2 act via a dominant negative mechanism with reduced penetrance and variable expressivity. Half of mutation carriers have no aortic disease. ACTA2 mutations (to date approximately 30 mutations have been identified) resulting in aortic aneurysms have been predominantly found to be missense (37), although deletions and splice-site mutations have also been described (36,38-44). Patients with ACTA2 mutations present with acute ascending (type A) or descending (type B) aortic dissections, and a median survival of 67 years (36). Although the aortic diameter is variable prior to rupture, most studies looking at ACTA2 mutations found the majority of aortic aneurysms to be <5.0 cm prior to dissection. For this reason, early surgical intervention should be considered even when minimal changes in aortic diameter are recognized. Histologically, ACTA2 mutations demonstrate vascular smooth muscle hyperplasia but not hypertrophy. This vascular smooth muscle hyperplasia has been associated with a possible increased risk of stroke and coronary artery disease (up to 25% in some studies) (38).
MYLK mutation
The MYLK gene is localized on chromosome 3 (3q21.1) and accounts for 1% of all FTAA mutations. Mutations in MYLK, which encodes myosin light chain kinase (MLCK), have been exclusively associated with thoracic ascending aortic dissections (TAAD). MLCK is a ubiquitously expressed kinase whose target of phosphorylation is a 20 kDa regulatory light chain (RLC) of myosin II (45). In smooth muscle cells initiation of myogenic response to mechanical stretch is a result of Ca/calmodulin complex activating MLCK. MLCK in turn phosphorylates RLC and this increases actin-activated myosin II ATPase and initiates the physiologic contraction of smooth muscle (37). MYLK mutations follow an autosomal dominant pattern of inheritance, and are thought to cause disease as a consequence of haplo-insufficiency. Because the pathophysiology of MYLK mutations is that of dissections and not aneurysms, it is difficult to counsel as to the time of intervention (37).
Conclusions
Medical science has made substantial progress in elucidating the genetic basis of aortic diseases since Marfan's original observations a century ago. The identification of specific mutations underlying syndromic and non-syndromic thoracic aortic aneurysms now permits precise identification of affected patients and confirmation of clinical diagnoses. Further, it is becoming clear that specific mutations lead to subtly different patterns of disease progression. Soon, we will enter an era of personalized aneurysm care, in which specific mutations will determine the appropriate size criterion for surgical intervention. Despite the great complexity of interpreting the wealth of information generated by whole genome sequencing, it is indeed this understanding that will allow aortic care to evolve beyond the current surgical plane, which, while delicate, intricate, and challenging, is essentially human “plumbing”.
Acknowledgements
Disclosure: This paper was originally presented at 5th International Congress on Aortic Surgery and Anesthesia: “How to do it”, in Milan in 2013, and published in the book for that course.
References
- Clouse WD, Hallett JW Jr, Schaff HV, et al. Acute aortic dissection: population-based incidence compared with degenerative aortic aneurysm rupture. Mayo Clin Proc 2004;79:176-80. [PubMed]
- El-Hamamsy I, Yacoub MH. Cellular and molecular mechanisms of thoracic aortic aneurysms. Nat Rev Cardiol 2009;6:771-86. [PubMed]
- Gott VL. Antoine Marfan and his syndrome: one hundred years later. Md Med J 1998;47:247-52. [PubMed]
- Loeys BL, Schwarze U, Holm T, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med 2006;355:788-98. [PubMed]
- Milewicz DM, Guo DC, Tran-Fadulu V, et al. Genetic basis of thoracic aortic aneurysms and dissections: focus on smooth muscle cell contractile dysfunction. Annu Rev Genomics Hum Genet 2008;9:283-302. [PubMed]
- Lucarini L, Evangelisti L, Attanasio M, et al. May TGFBR1 act also as low penetrance allele in Marfan syndrome? Int J Cardiol 2009;131:281-4. [PubMed]
- Bolar N, Van Laer L, Loeys BL. Marfan syndrome: from gene to therapy. Curr Opin Pediatr 2012;24:498-504. [PubMed]
- Loeys B, De Backer J, Van Acker P, et al. Comprehensive molecular screening of the FBN1 gene favors locus homogeneity of classical Marfan syndrome. Hum Mutat 2004;24:140-6. [PubMed]
- Halliday DJ, Hutchinson S, Lonie L, et al. Twelve novel FBN1 mutations in Marfan syndrome and Marfan related phenotypes test the feasibility of FBN1 mutation testing in clinical practice. J Med Genet 2002;39:589-93. [PubMed]
- Bunton TE, Biery NJ, Myers L, et al. Phenotypic alteration of vascular smooth muscle cells precedes elastolysis in a mouse model of Marfan syndrome. Circ Res 2001;88:37-43. [PubMed]
- Loeys BL, Chen J, Neptune ER, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet 2005;37:275-81. [PubMed]
- Van Hemelrijk C, Renard M, Loeys B. The Loeys-Dietz syndrome: an update for the clinician. Curr Opin Cardiol 2010;25:546-51. [PubMed]
- Jones JA, Spinale FG, Ikonomidis JS. Transforming growth factor-beta signaling in thoracic aortic aneurysm development: a paradox in pathogenesis. J Vasc Res 2009;46:119-37. [PubMed]
- Regalado ES, Guo DC, Villamizar C, et al. Exome sequencing identifies SMAD3 mutations as a cause of familial thoracic aortic aneurysm and dissection with intracranial and other arterial aneurysms. Circ Res 2011;109:680-6. [PubMed]
- van de Laar IM, van der Linde D, Oei EH, et al. Phenotypic spectrum of the SMAD3-related aneurysms-osteoarthritis syndrome. J Med Genet 2012;49:47-57. [PubMed]
- van de Laar IM, Oldenburg RA, Pals G, et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet 2011;43:121-6. [PubMed]
- van der Linde D, van de Laar IM, Bertoli-Avella AM, et al. Aggressive cardiovascular phenotype of aneurysms-osteoarthritis syndrome caused by pathogenic SMAD3 variants. J Am Coll Cardiol 2012;60:397-403. [PubMed]
- Coucke PJ, Willaert A, Wessels MW, et al. Mutations in the facilitative glucose transporter GLUT10 alter angiogenesis and cause arterial tortuosity syndrome. Nat Genet 2006;38:452-7. [PubMed]
- Coucke PJ, Wessels MW, Van Acker P, et al. Homozygosity mapping of a gene for arterial tortuosity syndrome to chromosome 20q13. J Med Genet 2003;40:747-51. [PubMed]
- Wessels MW, Catsman-Berrevoets CE, Mancini GM, et al. Three new families with arterial tortuosity syndrome. Am J Med Genet A 2004;131:134-43. [PubMed]
- Germain DP. Ehlers-Danlos syndrome type IV. Orphanet J Rare Dis 2007;2:32. [PubMed]
- Oderich GS, Panneton JM, Bower TC, et al. The spectrum, management and clinical outcome of Ehlers-Danlos syndrome type IV: a 30-year experience. J Vasc Surg 2005;42:98-106. [PubMed]
- Pepin M, Schwarze U, Superti-Furga A, et al. Clinical and genetic features of Ehlers-Danlos syndrome type IV, the vascular type. N Engl J Med 2000;342:673-80. [PubMed]
- Boileau C, Guo DC, Hanna N, et al. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat Genet 2012;44:916-21. [PubMed]
- Tilson MD, Dang C. Generalized arteriomegaly. A possible predisposition to the formation of abdominal aortic aneurysms. Arch Surg 1981;116:1030-2. [PubMed]
- Tilson MD, Seashore MR. Fifty families with abdominal aortic aneurysms in two or more first-order relatives. Am J Surg 1984;147:551-3. [PubMed]
- Biddinger A, Rocklin M, Coselli J, et al. Familial thoracic aortic dilatations and dissections: a case control study. J Vasc Surg 1997;25:506-11. [PubMed]
- Coady MA, Davies RR, Roberts M, et al. Familial patterns of thoracic aortic aneurysms. Arch Surg 1999;134:361-7. [PubMed]
- Milewicz DM, Carlson AA, Regalado ES. Genetic testing in aortic aneurysm disease: PRO. Cardiol Clin 2010;28:191-7. [PubMed]
- Albornoz G, Coady MA, Roberts M, et al. Familial thoracic aortic aneurysms and dissections--incidence, modes of inheritance, and phenotypic patterns. Ann Thorac Surg 2006;82:1400-5. [PubMed]
- Guo D, Hasham S, Kuang SQ, et al. Familial thoracic aortic aneurysms and dissections: genetic heterogeneity with a major locus mapping to 5q13-14. Circulation 2001;103:2461-8. [PubMed]
- Vaughan CJ, Casey M, He J, et al. Identification of a chromosome 11q23.2-q24 locus for familial aortic aneurysm disease, a genetically heterogeneous disorder. Circulation 2001;103:2469-75. [PubMed]
- Pannu H, Tran-Fadulu V, Papke CL, et al. MYH11 mutations result in a distinct vascular pathology driven by insulin-like growth factor 1 and angiotensin II. Hum Mol Genet 2007;16:2453-62. [PubMed]
- Zhu L, Vranckx R, Khau Van Kien P, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet 2006;38:343-9. [PubMed]
- Kuang SQ, Kwartler CS, Byanova KL, et al. Rare, nonsynonymous variant in the smooth muscle-specific isoform of myosin heavy chain, MYH11, R247C, alters force generation in the aorta and phenotype of smooth muscle cells. Circ Res 2012;110:1411-22. [PubMed]
- Guo DC, Pannu H, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet 2007;39:1488-93. [PubMed]
- Wang L, Guo DC, Cao J, et al. Mutations in myosin light chain kinase cause familial aortic dissections. Am J Hum Genet 2010;87:701-7. [PubMed]
- Guo DC, Papke CL, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am J Hum Genet 2009;84:617-27. [PubMed]
- Hoffjan S, Waldmüller S, Blankenfeldt W, et al. Three novel mutations in the ACTA2 gene in German patients with thoracic aortic aneurysms and dissections. Eur J Hum Genet 2011;19:520-4. [PubMed]
- Yoo EH, Choi SH, Jang SY, et al. Clinical, pathological, and genetic analysis of a Korean family with thoracic aortic aneurysms and dissections carrying a novel Asp26Tyr mutation. Ann Clin Lab Sci 2010;40:278-84. [PubMed]
- Disabella E, Grasso M, Gambarin FI, et al. Risk of dissection in thoracic aneurysms associated with mutations of smooth muscle alpha-actin 2 (ACTA2). Heart 2011;97:321-6. [PubMed]
- Renard M, Callewaert B, Baetens M, et al. Novel MYH11 and ACTA2 mutations reveal a role for enhanced TGFbeta signaling in FTAAD. Int J Cardiol 2013;165:314-21. [PubMed]
- Milewicz DM, Østergaard JR, Ala-Kokko LM, et al. De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am J Med Genet A 2010;152A:2437-43. [PubMed]
- Morisaki H, Akutsu K, Ogino H, et al. Mutation of ACTA2 gene as an important cause of familial and nonfamilial nonsyndromatic thoracic aortic aneurysm and/or dissection (TAAD). Hum Mutat 2009;30:1406-11. [PubMed]
- Kamm KE, Stull JT. Dedicated myosin light chain kinases with diverse cellular functions. J Biol Chem 2001;276:4527-30. [PubMed]





